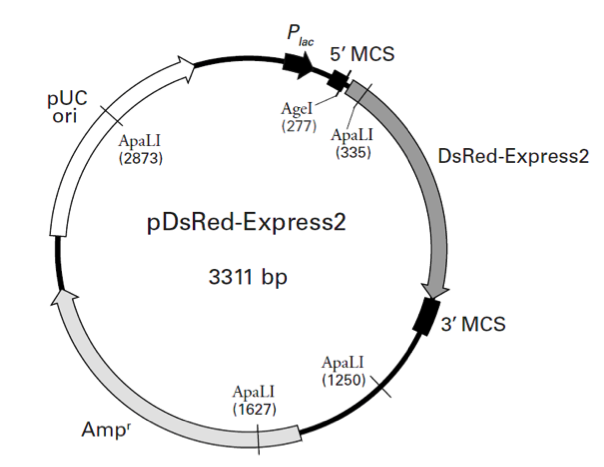
pDsRed-Express2
pDsRed-Express2
编号 | 载体名称 |
北京华越洋生物VECT4020 | pDsRed-Express2 |
pDsRed-Express2载体基本信息
载体名称: | pDsRed-Express2 |
质粒类型: | 大肠杆菌表达载体;cDNA克隆载体 |
高拷贝/低拷贝: | 高拷贝 |
克隆方法: | 限制性内切酶;多克隆位点 |
启动子: | lac |
载体大小: | 3311 bp |
5' 测序引物及序列: | -- |
3' 测序引物及序列: | -- |
载体标签: | DsRed-Express2 |
载体抗性: | 氨苄青霉素(Ampicillin) |
克隆菌株: | DH5alpha |
表达菌株: | 原核细胞 |
备注: | pDsRed-Express2载体是原核表达载体,也用作DsRed-Express2的cDNA克隆载体; |
稳定性: | 稳表达 |
组成型/诱导型: | -- |
病毒/非病毒: | 非病毒 |
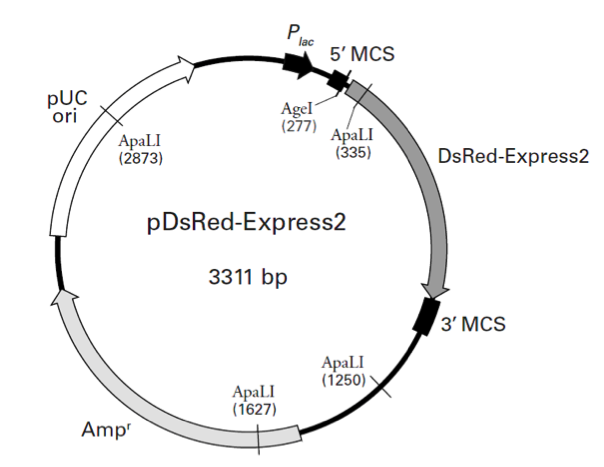
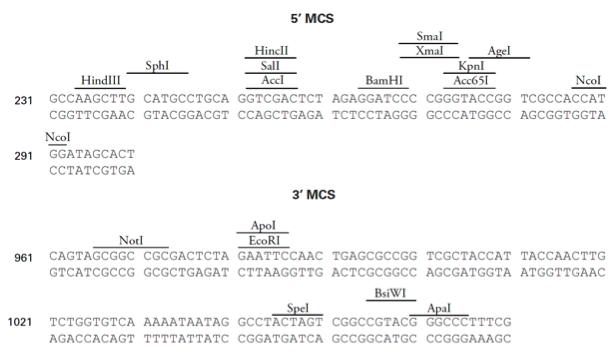
pDsRed-Express2载体质粒图谱和多克隆位点信息
pDsRed-Express2载体描述
pDsRed-Express2 is a prokaryotic expression vector that encodes DsRed-Express2, a variant of the Discosoma sp. red fluorescent protein, DsRed (1). DsRed-Express2 retains the fast maturation and high photostability characteristic of its predecessor, DsRed-Express (2), and has been engineered (through additional amino acid substitutions) for increased solubility and reduced cytotoxicity (3). Although it most likely forms the same tetrameric structure as wild-type DsRed, DsRed-Express2 displays a greatly reduced tendency to aggregate; this results in minimal cytotoxicity, which makes DsRed-Express2 much better suited for in vivo applications involving sensitive cells, such as primary or stem cells. DsRed-Express2 also exhibits extremely low residual green fluorescence, which allows cells expressing the protein to be effectively separated from other fluorescently labeled cell populations by flow cytometry.
In pDsRed-Express2, the DsRed-Express2 coding sequence is flanked by separate and distinct multiple cloning sites (i.e., the 5' MCS and 3' MCS) that make it easy to excise the gene for use in other cloning applications. In E. coli, DsRed-Express2 is expressed from the lac promoter (Plac) as a fusion with several amino acids, including the first five amino acids of the LacZ protein. Note, however, that if the DsRed-Express2 coding sequence is excised using a restriction site in the 5' MCS, the protein will no longer be expressed as a fusion (as it is when it is expressed from the lac promoter). A Kozak consensus sequence is located immediately upstream of the DsRed-Express2 coding sequence to enhance translational efficiency in eukaryotic cells (4). The entire DsRed-Express2 expression cassette in pDsRed-Express2 is supported by a pUC19 backbone, which contains a high-copy number origin of replication and an ampicillin resistance gene (Ampr) for propagation and selection in E. coli.
pDsRed-Express2 is primarily intended to serve as a source of DsRed-Express2 cDNA. The flanking MCS regions make it possible to excise the DsRed-Express2 coding sequence and insert it into other vector systems. The vector can also be used to express DsRed-Express2 in bacteria. Cells expressing DsRed-Express2 (excitation and emission maxima: 554 nm and 591 nm, respectively) can be detected by either fluorescence microscopy or flow cytometry 8–12 hours after transfection.
For Western analysis, DsRed-Express2 can be detected with either the Living Colors DsRed Polyclonal Anti-body or the Living Colors DsRed Monoclonal Antibody.
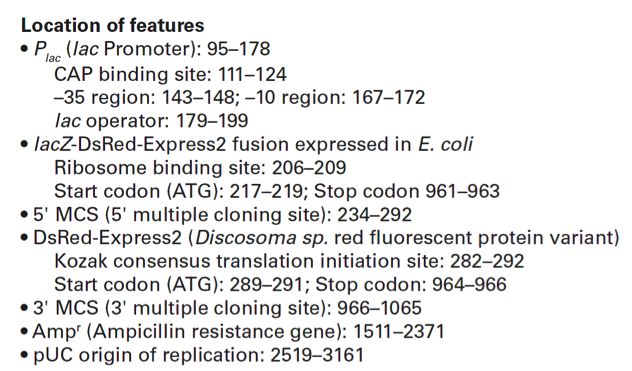
pDsRed-Express2载体含有以下元件:
Plac (lac Promoter): 95–178
CAP binding site: 111–124
–35 region: 143–148; –10 region: 167–172
lac operator: 179–199
lacZ-DsRed-Express2 fusion expressed in E. coli
Ribosome binding site: 206–209
Start codon (ATG): 217–219; Stop codon 961–963
5' MCS (5' multiple cloning site): 234–292
DsRed-Express2 (Discosoma sp. red fluorescent protein variant)
Kozak consensus translation initiation site: 282–292
Start codon (ATG): 289–291; Stop codon: 964–966
3' MCS (3' multiple cloning site): 966–1065
Ampr (Ampicillin resistance gene): 1511–2371
pUC origin of replication: 2519–3161
Propagation in E. coli
Recommended host strain: DH5α
Selectable marker: plasmid confers resistance to ampicillin (50 μg/ml) in E. coli hosts.
E. coli replication origin: pUC
Copy number: high
Plasmid incompatibility group: pMB1/ColE1
Excitation and emission maxima of DsRed-Express2
Excitation maximum = 554 nm
Emission maximum = 591 nm
载体序列
ORIGIN
1 AGCGCCCAAT ACGCAAACCG CCTCTCCCCG CGCGTTGGCC GATTCATTAA TGCAGCTGGC
61 ACGACAGGTT TCCCGACTGG AAAGCGGGCA GTGAGCGCAA CGCAATTAAT GTGAGTTAGC
121 TCACTCATTA GGCACCCCAG GCTTTACACT TTATGCTTCC GGCTCGTATG TTGTGTGGAA
181 TTGTGAGCGG ATAACAATTT CACACAGGAA ACAGCTATGA CCATGATTAC GCCAAGCTTG
241 CATGCCTGCA GGTCGACTCT AGAGGATCCC CGGGTACCGG TCGCCACCAT GGATAGCACT
301 GAGAACGTCA TCAAGCCCTT CATGCGCTTC AAGGTGCACA TGGAGGGCTC CGTGAACGGC
361 CACGAGTTCG AGATCGAGGG CGAGGGCGAG GGCAAGCCCT ACGAGGGCAC CCAGACCGCC
421 AAGCTGCAGG TGACCAAGGG CGGCCCCCTG CCCTTCGCCT GGGACATCCT GTCCCCCCAG
481 TTCCAGTACG GCTCCAAGGT GTACGTGAAG CACCCCGCCG ACATCCCCGA CTACAAGAAG
541 CTGTCCTTCC CCGAGGGCTT CAAGTGGGAG CGCGTGATGA ACTTCGAGGA CGGCGGCGTG
601 GTGACCGTGA CCCAGGACTC CTCCCTGCAG GACGGCACCT TCATCTACCA CGTGAAGTTC
661 ATCGGCGTGA ACTTCCCCTC CGACGGCCCC GTAATGCAGA AGAAGACTCT GGGCTGGGAG
721 CCCTCCACCG AGCGCCTGTA CCCCCGCGAC GGCGTGCTGA AGGGCGAGAT CCACAAGGCG
781 CTGAAGCTGA AGGGCGGCGG CCACTACCTG GTGGAGTTCA AGTCAATCTA CATGGCCAAG
841 AAGCCCGTGA AGCTGCCCGG CTACTACTAC GTGGACTCCA AGCTGGACAT CACCTCCCAC
901 AACGAGGACT ACACCGTGGT GGAGCAGTAC GAGCGCGCCG AGGCCCGCCA CCACCTGTTC
961 CAGTAGCGGC CGCGACTCTA GAATTCCAAC TGAGCGCCGG TCGCTACCAT TACCAACTTG
1021 TCTGGTGTCA AAAATAATAG GCCTACTAGT CGGCCGTACG GGCCCTTTCG TCTCGCGCGT
1081 TTCGGTGATG ACGGTGAAAA CCTCTGACAC ATGCAGCTCC CGGAGACGGT CACAGCTTGT
1141 CTGTAAGCGG ATGCCGGGAG CAGACAAGCC CGTCAGGGCG CGTCAGCGGG TGTTGGCGGG
1201 TGTCGGGGCT GGCTTAACTA TGCGGCATCA GAGCAGATTG TACTGAGAGT GCACCATATG
1261 CGGTGTGAAA TACCGCACAG ATGCGTAAGG AGAAAATACC GCATCAGGCG GCCTTAAGGG
1321 CCTCGTGATA CGCCTATTTT TATAGGTTAA TGTCATGATA ATAATGGTTT CTTAGACGTC
1381 AGGTGGCACT TTTCGGGGAA ATGTGCGCGG AACCCCTATT TGTTTATTTT TCTAAATACA
1441 TTCAAATATG TATCCGCTCA TGAGACAATA ACCCTGATAA ATGCTTCAAT AATATTGAAA
1501 AAGGAAGAGT ATGAGTATTC AACATTTCCG TGTCGCCCTT ATTCCCTTTT TTGCGGCATT
1561 TTGCCTTCCT GTTTTTGCTC ACCCAGAAAC GCTGGTGAAA GTAAAAGATG CTGAAGATCA
1621 GTTGGGTGCA CGAGTGGGTT ACATCGAACT GGATCTCAAC AGCGGTAAGA TCCTTGAGAG
1681 TTTTCGCCCC GAAGAACGTT TTCCAATGAT GAGCACTTTT AAAGTTCTGC TATGTGGCGC
1741 GGTATTATCC CGTATTGACG CCGGGCAAGA GCAACTCGGT CGCCGCATAC ACTATTCTCA
1801 GAATGACTTG GTTGAGTACT CACCAGTCAC AGAAAAGCAT CTTACGGATG GCATGACAGT
1861 AAGAGAATTA TGCAGTGCTG CCATAACCAT GAGTGATAAC ACTGCGGCCA ACTTACTTCT
1921 GACAACGATC GGAGGACCGA AGGAGCTAAC CGCTTTTTTG CACAACATGG GGGATCATGT
1981 AACTCGCCTT GATCGTTGGG AACCGGAGCT GAATGAAGCC ATACCAAACG ACGAGCGTGA
2041 CACCACGATG CCTGTAGCAA TGGCAACAAC GTTGCGCAAA CTATTAACTG GCGAACTACT
2101 TACTCTAGCT TCCCGGCAAC AATTAATAGA CTGGATGGAG GCGGATAAAG TTGCAGGACC
2161 ACTTCTGCGC TCGGCCCTTC CGGCTGGCTG GTTTATTGCT GATAAATCTG GAGCCGGTGA
2221 GCGTGGGTCT CGCGGTATCA TTGCAGCACT GGGGCCAGAT GGTAAGCCCT CCCGTATCGT
2281 AGTTATCTAC ACGACGGGGA GTCAGGCAAC TATGGATGAA CGAAATAGAC AGATCGCTGA
2341 GATAGGTGCC TCACTGATTA AGCATTGGTA ACTGTCAGAC CAAGTTTACT CATATATACT
2401 TTAGATTGAT TTAAAACTTC ATTTTTAATT TAAAAGGATC TAGGTGAAGA TCCTTTTTGA
2461 TAATCTCATG ACCAAAATCC CTTAACGTGA GTTTTCGTTC CACTGAGCGT CAGACCCCGT
2521 AGAAAAGATC AAAGGATCTT CTTGAGATCC TTTTTTTCTG CGCGTAATCT GCTGCTTGCA
2581 AACAAAAAAA CCACCGCTAC CAGCGGTGGT TTGTTTGCCG GATCAAGAGC TACCAACTCT
2641 TTTTCCGAAG GTAACTGGCT TCAGCAGAGC GCAGATACCA AATACTGTTC TTCTAGTGTA
2701 GCCGTAGTTA GGCCACCACT TCAAGAACTC TGTAGCACCG CCTACATACC TCGCTCTGCT
2761 AATCCTGTTA CCAGTGGCTG CTGCCAGTGG CGATAAGTCG TGTCTTACCG GGTTGGACTC
2821 AAGACGATAG TTACCGGATA AGGCGCAGCG GTCGGGCTGA ACGGGGGGTT CGTGCACACA
2881 GCCCAGCTTG GAGCGAACGA CCTACACCGA ACTGAGATAC CTACAGCGTG AGCTATGAGA
2941 AAGCGCCACG CTTCCCGAAG GGAGAAAGGC GGACAGGTAT CCGGTAAGCG GCAGGGTCGG
3001 AACAGGAGAG CGCACGAGGG AGCTTCCAGG GGGAAACGCC TGGTATCTTT ATAGTCCTGT
3061 CGGGTTTCGC CACCTCTGAC TTGAGCGTCG ATTTTTGTGA TGCTCGTCAG GGGGGCGGAG
3121 CCTATGGAAA AACGCCAGCA ACGCGGCCTT TTTACGGTTC CTGGCCTTTT GCTGGCCTTT
3181 TGCTCACATG TTCTTTCCTG CGTTATCCCC TGATTCTGTG GATAACCGTA TTACCGCCTT
3241 TGAGTGAGCT GATACCGCTC GCCGCAGCCG AACGACCGAG CGCAGCGAGT CAGTGAGCGA
3301 GGAAGCGGAA G
//
pDsRed-Express2其他大肠杆菌表达载体: