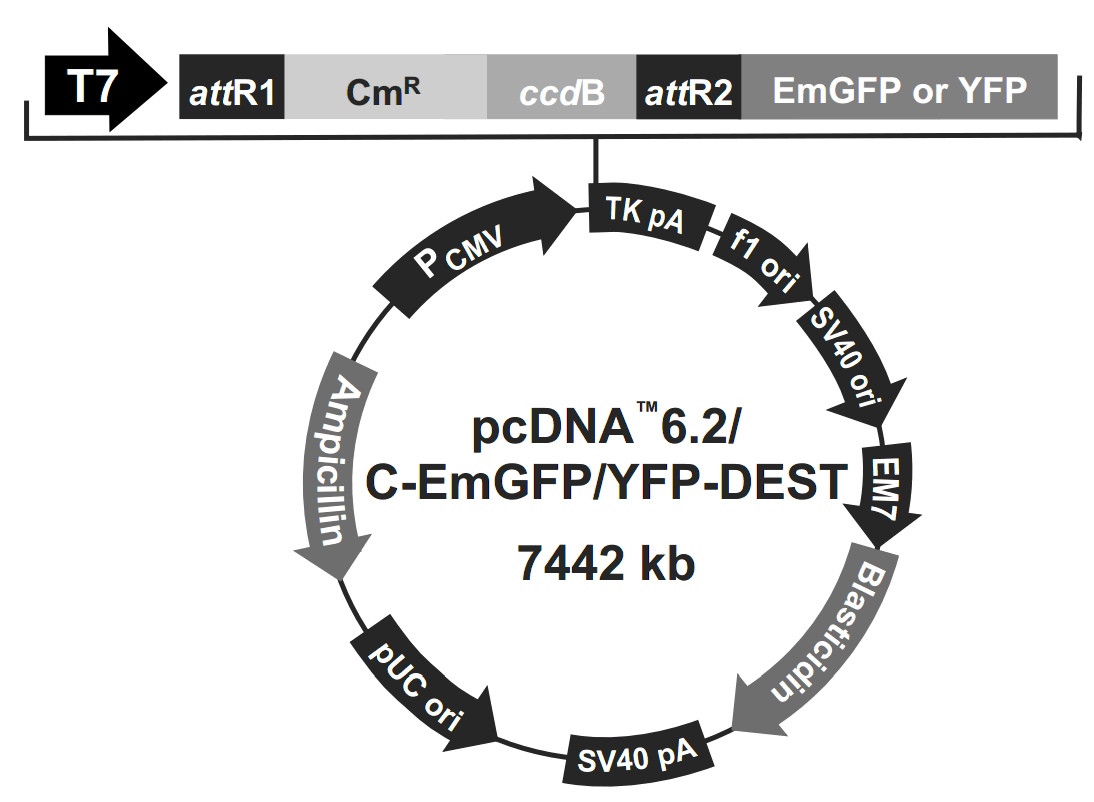
pcDNA6.2/C-EmGFP-DEST
pcDNA6.2/C-EmGFP-DEST
编号 | 载体名称 |
北京华越洋VECT511203 | pcDNA6.2/C-EmGFP-DEST |
pcDNA6.2/C-EmGFP-DEST载体基本信息:
载体名称: | pcDNA6.2/C-EmGFP-DEST |
质粒类型: | 哺乳动物表达载体;Gateway载体;荧光报告载体 |
高拷贝/低拷贝: | 高拷贝 |
启动子: | CMV |
克隆方法: | Gateway |
载体大小: | 7711bp |
5' 测序引物及序列: | T7 forward:5’-TAATACGACTCACTATAGGG-3' |
3' 测序引物及序列: | TK polyA Reverse: 5’-CTTCCGTGTTTCAGTTAGC-3’ |
载体标签: | EmGFP Tag (C Term) |
载体抗性: | 氨苄青霉素、氯霉素(仅空载体具有此抗性) |
筛选标记: | Blasticidin |
克隆菌株: | ccdB阳性筛选 DB3.1 感受态细胞, 阴性筛选用OmniMAX2-T1感受态细胞 |
表达细胞(系): | 常规细胞系,如293、Hela等。 |
备注: |
pcDNA6.2/C-EmGFP-DEST载体是cDNA表达载体,是绿色荧光报告载体; |
稳定性: | 瞬表达 或 稳表达 |
组成型/诱导型: | 组成型 |
病毒/非病毒: | 非病毒 |
pcDNA6.2/C-EmGFP-DEST载体质粒图谱和多克隆位点信息:

pcDNA6.2/C-EmGFP-DEST载体简介:
系统介绍
The Vivid Colors Fluorescent Protein Gateway Destination Vectors allow you to quickly and easily fuse a protein of interest to the widely used and well-characterized fluorescent proteinsfrom the jellyfish Aequorea victoria (1,2) using the pcDNA 6.2 Gateway mammalian expression vector. These powerful Gateway Technology vectors contain the next-generation EGFP, Emerald Green Fluorescent Protein (EmGFP), or the popular Yellow Fluorescent Protein (YFP) for simple, non-invasive detection of recombinant protein. Both fluorescent proteins have been humanizedfor optimal mammalian expression (3). In addition, the Vivid Colors pcDNA 6.2 Fluorescent Protein Gateway Vectors offer:
CMV promoter for high-level expression of the recombinant fluorescent fusion protein
Options to fuse EmGFP or YFP to the N- and C-terminus of your protein
Vivid Colors pcDNA6.2/EmGFP and YFP-DEST vectors combine the ease and flexibility of Gateway recombination based cloning with the brightness of Emerald Green Fluorescent Protein (EmGFP) and Yellow Fluorescent Protein (YFP) derived from Aequorea victoria GFP. Users can easily make an EmGFP or YFP N- or C-terminally tagged mammalian expression clone by performing an LR recombination reaction between a Gateway entry vector containing the gene of choice and a Vivid Colors pcDNA6.2/EmGFP or YFP-DEST Gateway Vector. After transfection of the expression clone into mammalian cells, the fluorescent-tagged protein of interest can be identified by fluorescence detection methods for localization experiments. The protein of interest can also be analyzed by Western blot.
The Vivid Colors pcDNA6.2/EmGFP or YFP-DEST Gateway Vectors are supplied with either an N- or C-terminal tagged EmGFP or YFP/GW/CAT plasmid that serves as a control for transfection efficiency of the expression clone into the target cells, as well as a control for expression of the gene of interest.
载体特征
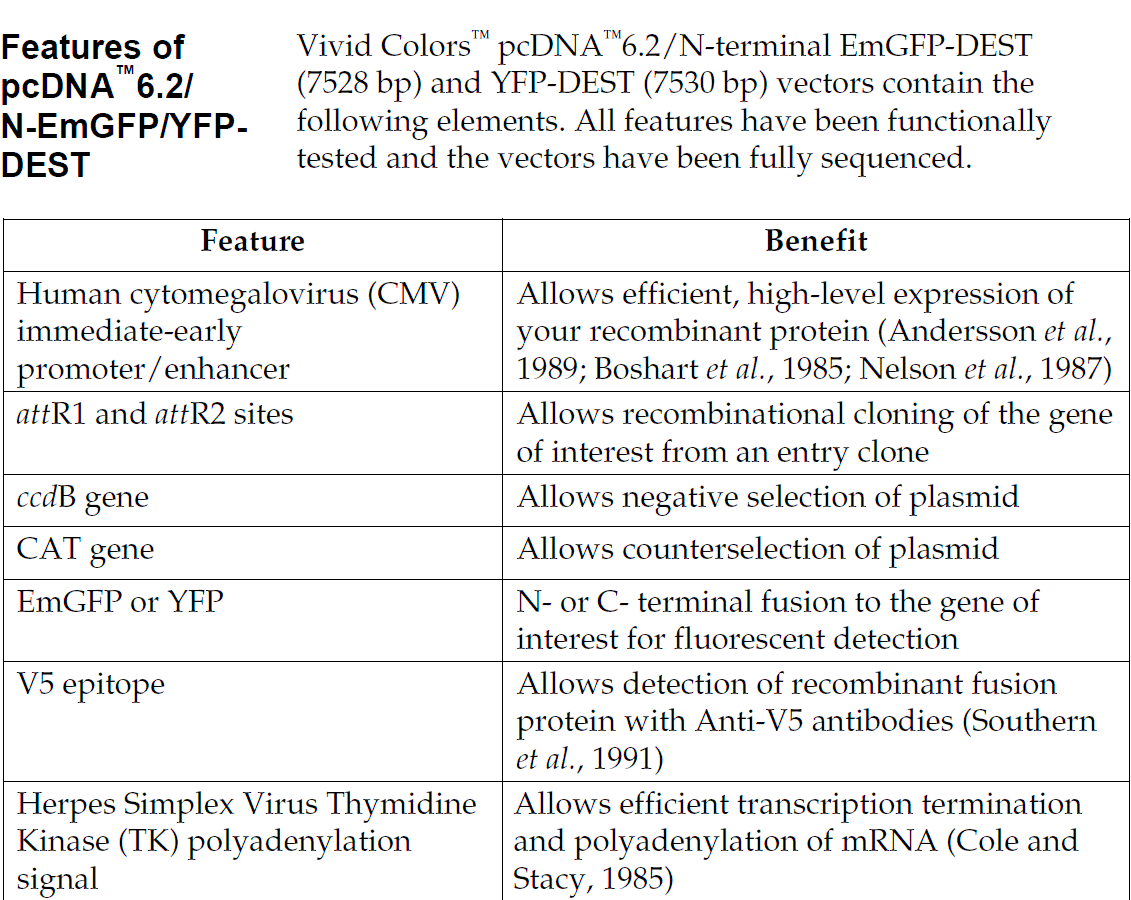
pcDNA6.2/EmGFP-DEST载体含有以下元件:
Human cytomegalovirus immediate-early (CMV) promoter/enhancer for high-level expression in a wide range of mammalian cells
Two recombination sites, attR1 and attR2, downstream of the CMV promoter for recombinational cloning of the gene of interest from an entry clone
Emerald Green Fluorescent Protein (EmGFP) or Yellow Fluorescent Protein (YFP) derived from Aequorea victoria GFP for N- or C-terminal fusion to the gene of interest for fluorescent detection
The V5 epitope tag for detection of recombinant protein using Anti-V5 antibodies (optional on N-terminal fusion vectors only)
CAT gene located between the two attR sites for counterselection
The ccdB gene located between the two attR sites for negative selection
The Herpes Simplex Virus thymidine kinase polyadenylation signal for proper termination and processing of the recombinant transcript
f1 intergenic region for production of single-strand DNA in F plasmid-containing E. coli
SV40 early promoter and origin for expression of the Blasticidin resistance gene and stable propagation of the plasmid in mammalian hosts expressing the SV40 large T antigen
Blasticidin resistance gene for selection of stable cell lines
The pUC origin for high copy replication and maintenance of the plasmid in E.coli
The ampicillin (bla) resistance gene for selection in E. coli
绿色荧光蛋白(GFP)
Green Fluorescent Protein (GFP) is a naturally occurring bioluminescent protein derived from the jellyfish Aequorea victoria (Shimomura et al., 1962). GFP emits fluorescence upon excitation, and the gene encoding GFP contains all of the necessary information for posttranslational synthesis of the luminescent protein. GFP is often used as a molecular beacon because it requires no species-specific cofactors for function, and the fluorescence is easily detected using fluorescence microscopy and standard filter sets. Commonly, GFP is fused to a protein of interest, and upon expression, the localization of the fusion protein can be detected in cells. GFP can also function as a reporter gene downstream of a promoter of interest.
Modifications have been made to the wild-type GFP to enhance its expression in mammalian systems. These modifications include amino acid substitutions that correspond to the codon preference for mammalian use, and mutations that increase the brightness of the fluorescence signal, resulting in “enhanced” GFP (Zhang et al., 1996).
Mutations have also arisen or have been introduced into GFP that further enhance and shift the spectral properties of GFP such that these proteins will emit fluorescent color variations (reviewed in Tsien, 1998). The Emerald GFP (EmGFP) and Yellow Fluorescent Protein (YFP) are such variants of enhanced GFP.
LR重组反应所需实验材料:
Purified plasmid DNA of your entry clone
(50-150 ng/μL in TE, pH 8.0)
Vivid Colors pcDNA6.2/EmGFP or YFP-DEST Gateway Vector (150 ng/μL in TE, pH 8.0)
LR Clonase II enzyme mix (page 34; keep at –20°C until immediately before use)
pENTR-gus (supplied with LR Clonase II enzyme mix; use as a control for the LR reaction; 50 ng/μL)
TE Buffer, pH 8.0 (10 mM Tris-HCl, pH 8.0, 1 mM EDTA)
2 μg/μL Proteinase K solution (supplied with LR Clonase II enzyme mix; thaw and keep on ice until use)
Appropriate competent E. coli host and growth medium for expression
S.O.C. Medium
LB agar plates containing 100 μg/mL ampicillin
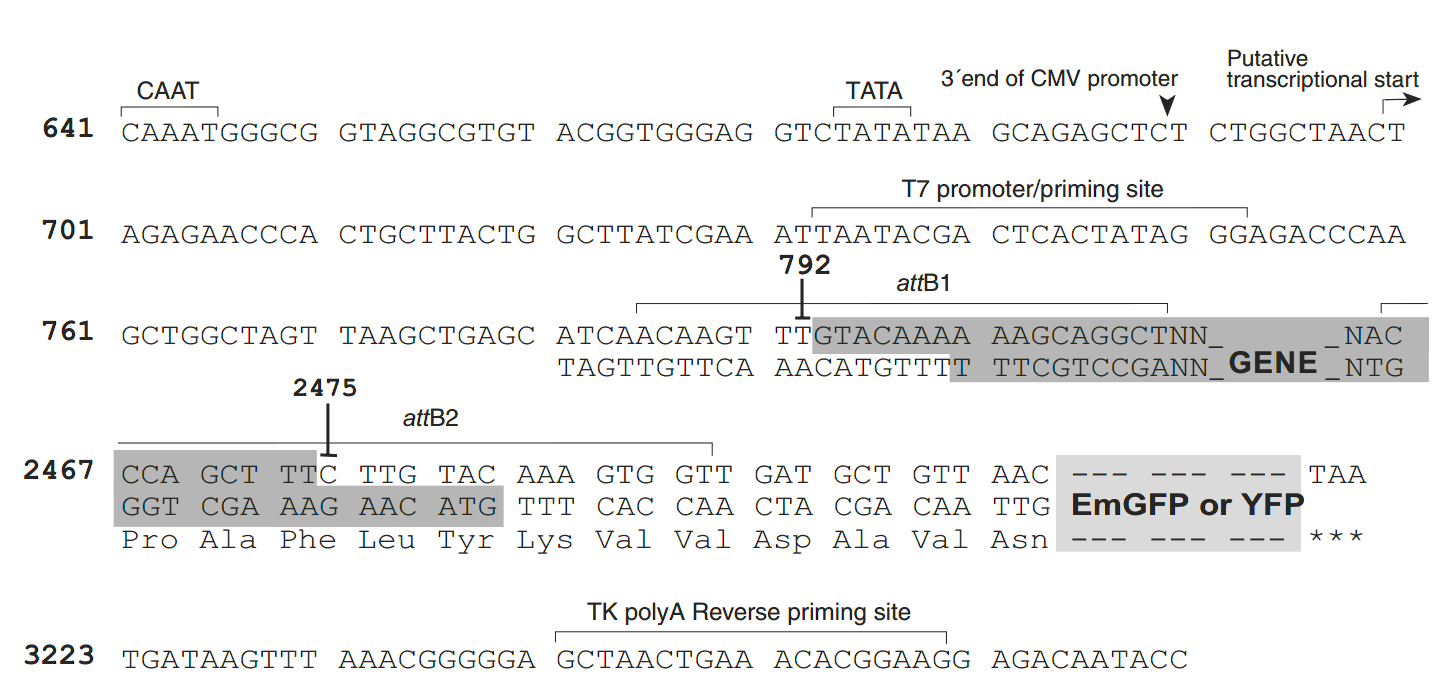
pcDNA6.2/C-EmGFP-DEST载体序列:
TTAAGCTACAACAAGGCAAGGCTTGACCGACAATTGCATGAAGAATCTGCTTAGGGTTAGGCGTTTTGCG
CTGCTTCGCGATGTACGGGCCAGATATACGCGTTGACATTGATTATTGACTAGTTATTAATAGTAATCAA
TTACGGGGTCATTAGTTCATAGCCCATATATGGAGTTCCGCGTTACATAACTTACGGTAAATGGCCCGCC
TGGCTGACCGCCCAACGACCCCCGCCCATTGACGTCAATAATGACGTATGTTCCCATAGTAACGCCAATA
GGGACTTTCCATTGACGTCAATGGGTGGAGTATTTACGGTAAACTGCCCACTTGGCAGTACATCAAGTGT
ATCATATGCCAAGTACGCCCCCTATTGACGTCAATGACGGTAAATGGCCCGCCTGGCATTATGCCCAGTA
CATGACCTTATGGGACTTTCCTACTTGGCAGTACATCTACGTATTAGTCATCGCTATTACCATGGTGATG
CGGTTTTGGCAGTACATCAATGGGCGTGGATAGCGGTTTGACTCACGGGGATTTCCAAGTCTCCACCCCA
TTGACGTCAATGGGAGTTTGTTTTGGCACCAAAATCAACGGGACTTTCCAAAATGTCGTAACAACTCCGC
CCCATTGACGCAAATGGGCGGTAGGCGTGTACGGTGGGAGGTCTATATAAGCAGAGCTCTCTGGCTAACT
AGAGAACCCACTGCTTACTGGCTTATCGAAATTAATACGACTCACTATAGGGAGACCCAAGCTGGCTAGT
TAAGCTGAGCATCAACAAGTTTGTACAAAAAAGCTGAACGAGAAACGTAAAATGATATAAATATCAATAT
ATTAAATTAGATTTTGCATAAAAAACAGACTACATAATACTGTAAAACACAACATATCCAGTCACTATGG
CGGCCGCATTAGGCACCCCAGGCTTTACACTTTATGCTTCCGGCTCGTATAATGTGTGGATTTTGAGTTA
GGATCCGTCGAGATTTTCAGGAGCTAAGGAAGCTAAAATGGAGAAAAAAATCACTGGATATACCACCGTT
GATATATCCCAATGGCATCGTAAAGAACATTTTGAGGCATTTCAGTCAGTTGCTCAATGTACCTATAACC
AGACCGTTCAGCTGGATATTACGGCCTTTTTAAAGACCGTAAAGAAAAATAAGCACAAGTTTTATCCGGC
CTTTATTCACATTCTTGCCCGCCTGATGAATGCTCATCCGGAATTCCGTATGGCAATGAAAGACGGTGAG
CTGGTGATATGGGATAGTGTTCACCCTTGTTACACCGTTTTCCATGAGCAAACTGAAACGTTTTCATCGC
TCTGGAGTGAATACCACGACGATTTCCGGCAGTTTCTACACATATATTCGCAAGATGTGGCGTGTTACGG
TGAAAACCTGGCCTATTTCCCTAAAGGGTTTATTGAGAATATGTTTTTCGTCTCAGCCAATCCCTGGGTG
AGTTTCACCAGTTTTGATTTAAACGTGGCCAATATGGACAACTTCTTCGCCCCCGTTTTCACCATGGGCA
AATATTATACGCAAGGCGACAAGGTGCTGATGCCGCTGGCGATTCAGGTTCATCATGCCGTTTGTGATGG
CTTCCATGTCGGCAGAATGCTTAATGAATTACAACAGTACTGCGATGAGTGGCAGGGCGGGGCGTAAAGA
TCTGGATCCGGCTTACTAAAAGCCAGATAACAGTATGCGTATTTGCGCGCTGATTTTTGCGGTATAAGAA
TATATACTGATATGTATACCCGAAGTATGTCAAAAAGAGGTATGCTATGAAGCAGCGTATTACAGTGACA
GTTGACAGCGACAGCTATCAGTTGCTCAAGGCATATATGATGTCAATATCTCCGGTCTGGTAAGCACAAC
CATGCAGAATGAAGCCCGTCGTCTGCGTGCCGAACGCTGGAAAGCGGAAAATCAGGAAGGGATGGCTGAG
GTCGCCCGGTTTATTGAAATGAACGGCTCTTTTGCTGACGAGAACAGGGGCTGGTGAAATGCAGTTTAAG
GTTTACACCTATAAAAGAGAGAGCCGTTATCGTCTGTTTGTGGATGTACAGAGTGATATTATTGACACGC
CCGGGCGACGGATGGTGATCCCCCTGGCCAGTGCACGTCTGCTGTCAGATAAAGTCTCCCGTGAACTTTA
CCCGGTGGTGCATATCGGGGATGAAAGCTGGCGCATGATGACCACCGATATGGCCAGTGTGCCGGTCTCC
GTTATCGGGGAAGAAGTGGCTGATCTCAGCCACCGCGAAAATGACATCAAAAACGCCATTAACCTGATGT
TCTGGGGAATATAAATGTCAGGCTCCCTTATACACAGCCAGTCTGCAGGTCGACCATAGTGACTGGATAT
GTTGTGTTTTACAGTATTATGTAGTCTGTTTTTTATGCAAAATCTAATTTAATATATTGATATTTATATC
ATTTTACGTTTCTCGTTCAGCTTTCTTGTACAAAGTGGTTGATGCTGTTAACATGGTGAGCAAGGGCGAG
GAGCTGTTCACCGGGGTGGTGCCCATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCG
TGTCCGGCGAGGGCGAGGGCGATGCCACCTACGGCAAGCTGACCCTGAAGTTCATCTGCACCACCGGCAA
GCTGCCCGTGCCCTGGCCCACCCTCGTGACCACCTTCACCTACGGCGTGCAGTGCTTCGCCCGCTACCCC
GACCACATGAAGCAGCACGACTTCTTCAAGTCCGCCATGCCCGAAGGCTACGTCCAGGAGCGCACCATCT
TCTTCAAGGACGACGGCAACTACAAGACCCGCGCCGAGGTGAAGTTCGAGGGCGACACCCTGGTGAACCG
CATCGAGCTGAAGGGCATCGACTTCAAGGAGGACGGCAACATCCTGGGGCACAAGCTGGAGTACAACTAC
AACAGCCACAAGGTCTATATCACCGCCGACAAGCAGAAGAACGGCATCAAGGTGAACTTCAAGACCCGCC
ACAACATCGAGGACGGCAGCGTGCAGCTCGCCGACCACTACCAGCAGAACACCCCCATCGGCGACGGCCC
CGTGCTGCTGCCCGACAACCACTACCTGAGCACCCAGTCCGCCCTGAGCAAAGACCCCAACGAGAAGCGC
GATCACATGGTCCTGCTGGAGTTCGTGACCGCCGCCGGGATCACTCTCGGCATGGACGAGCTGTACAAGT
AATGATAAGTTTAAACGGGGGAGGCTAACTGAAACACGGAAGGAGACAATACCGGAAGGAACCCGCGCTA
TGACGGCAATAAAAAGACAGAATAAAACGCACGGGTGTTGGGTCGTTTGTTCATAAACGCGGGGTTCGGT
CCCAGGGCTGGCACTCTGTCGATACCCCACCGAGACCCCATTGGGGCCAATACGCCCGCGTTTCTTCCTT
TTCCCCACCCCACCCCCCAAGTTCGGGTGAAGGCCCAGGGCTCGCAGCCAACGTCGGGGCGGCAGGCCCT
GCCATAGCAGATCTGCGCAGCTGGGGCTCTAGGGGGTATCCCCACGCGCCCTGTAGCGGCGCATTAAGCG
CGGCGGGTGTGGTGGTTACGCGCAGCGTGACCGCTACACTTGCCAGCGCCCTAGCGCCCGCTCCTTTCGC
TTTCTTCCCTTCCTTTCTCGCCACGTTCGCCGGCTTTCCCCGTCAAGCTCTAAATCGGGGGCTCCCTTTA
GGGTTCCGATTTAGTGCTTTACGGCACCTCGACCCCAAAAAACTTGATTAGGGTGATGGTTCACGTAGTG
GGCCATCGCCCTGATAGACGGTTTTTCGCCCTTTGACGTTGGAGTCCACGTTCTTTAATAGTGGACTCTT
GTTCCAAACTGGAACAACACTCAACCCTATCTCGGTCTATTCTTTTGATTTATAAGGGATTTTGCCGATT
TCGGCCTATTGGTTAAAAAATGAGCTGATTTAACAAAAATTTAACGCGAATTAATTCTGTGGAATGTGTG
TCAGTTAGGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAG
TCAGCAACCAGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATT
AGTCAGCAACCATAGTCCCGCCCCTAACTCCGCCCATCCCGCCCCTAACTCCGCCCAGTTCCGCCCATTC
TCCGCCCCATGGCTGACTAATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCTGCCTCTGAGCTATTC
CAGAAGTAGTGAGGAGGCTTTTTTGGAGGCCTAGGCTTTTGCAAAAAGCTCCCGGGAGCTTGTATATCCA
TTTTCGGATCTGATCAGCACGTGTTGACAATTAATCATCGGCATAGTATATCGGCATAGTATAATACGAC
AAGGTGAGGAACTAAACCATGGCCAAGCCTTTGTCTCAAGAAGAATCCACCCTCATTGAAAGAGCAACGG
CTACAATCAACAGCATCCCCATCTCTGAAGACTACAGCGTCGCCAGCGCAGCTCTCTCTAGCGACGGCCG
CATCTTCACTGGTGTCAATGTATATCATTTTACTGGGGGACCTTGTGCAGAACTCGTGGTGCTGGGCACT
GCTGCTGCTGCGGCAGCTGGCAACCTGACTTGTATCGTCGCGATCGGAAATGAGAACAGGGGCATCTTGA
GCCCCTGCGGACGGTGCCGACAGGTGCTTCTCGATCTGCATCCTGGGATCAAAGCCATAGTGAAGGACAG
TGATGGACAGCCGACGGCAGTTGGGATTCGTGAATTGCTGCCCTCTGGTTATGTGTGGGAGGGCTAAGCA
CTTCGTGGCCGAGGAGCAGGACTGACACGTGCTACGAGATTTCGATTCCACCGCCGCCTTCTATGAAAGG
TTGGGCTTCGGAATCGTTTTCCGGGACGCCGGCTGGATGATCCTCCAGCGCGGGGATCTCATGCTGGAGT
TCTTCGCCCACCCCAACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCACAAATTT
CACAAATAAAGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTCCAAACTCATCAATGTATCTTATCAT
GTCTGTATACCGTCGACCTCTAGCTAGAGCTTGGCGTAATCATGGTCATAGCTGTTTCCTGTGTGAAATT
GTTATCCGCTCACAATTCCACACAACATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATG
AGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAG
CTGCATTAATGAATCGGCCAACGCGCGGGGAGAGGCGGTTTGCGTATTGGGCGCTCTTCCGCTTCCTCGC
TCACTGACTCGCTGCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAAAGGCGGTAATACG
GTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAAC
CGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGAC
GCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCT
CGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTG
GCGCTTTCTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTG
TGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATCGTCTTGAGTCCAACCCGGT
AAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGT
GCTACAGAGTTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGAACAGTATTTGGTATCTGCGCTC
TGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAG
CGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATC
TTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGAGATTATCAA
AAAGGATCTTCACCTAGATCCTTTTAAATTAAAAATGAAGTTTTAAATCAATCTAAAGTATATATGAGTA
AACTTGGTCTGACAGTTACCAATGCTTAATCAGTGAGGCACCTATCTCAGCGATCTGTCTATTTCGTTCA
TCCATAGTTGCCTGACTCCCCGTCGTGTAGATAACTACGATACGGGAGGGCTTACCATCTGGCCCCAGTG
CTGCAATGATACCGCGAGACCCACGCTCACCGGCTCCAGATTTATCAGCAATAAACCAGCCAGCCGGAAG
GGCCGAGCGCAGAAGTGGTCCTGCAACTTTATCCGCCTCCATCCAGTCTATTAATTGTTGCCGGGAAGCT
AGAGTAAGTAGTTCGCCAGTTAATAGTTTGCGCAACGTTGTTGCCATTGCTACAGGCATCGTGGTGTCAC
GCTCGTCGTTTGGTATGGCTTCATTCAGCTCCGGTTCCCAACGATCAAGGCGAGTTACATGATCCCCCAT
GTTGTGCAAAAAAGCGGTTAGCTCCTTCGGTCCTCCGATCGTTGTCAGAAGTAAGTTGGCCGCAGTGTTA
TCACTCATGGTTATGGCAGCACTGCATAATTCTCTTACTGTCATGCCATCCGTAAGATGCTTTTCTGTGA
CTGGTGAGTACTCAACCAAGTCATTCTGAGAATAGTGTATGCGGCGACCGAGTTGCTCTTGCCCGGCGTC
AATACGGGATAATACCGCGCCACATAGCAGAACTTTAAAAGTGCTCATCATTGGAAAACGTTCTTCGGGG
CGAAAACTCTCAAGGATCTTACCGCTGTTGAGATCCAGTTCGATGTAACCCACTCGTGCACCCAACTGAT
CTTCAGCATCTTTTACTTTCACCAGCGTTTCTGGGTGAGCAAAAACAGGAAGGCAAAATGCCGCAAAAAA
GGGAATAAGGGCGACACGGAAATGTTGAATACTCATACTCTTCCTTTTTCAATATTATTGAAGCATTTAT
CAGGGTTATTGTCTCATGAGCGGATACATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGC
GCACATTTCCCCGAAAAGTGCCACCTGACGTCGACGGATCGGGAGATCTCCCGATCCCCTATGGTGCACT
CTCAGTACAATCTGCTCTGATGCCGCATAGTTAAGCCAGTATCTGCTCCCTGCTTGTGTGTTGGAGGTCG
CTGAGTAGTGCGCGAGCAAAAT
pcDNA6.2/C-EmGFP-DEST其他相关Gateway载体: