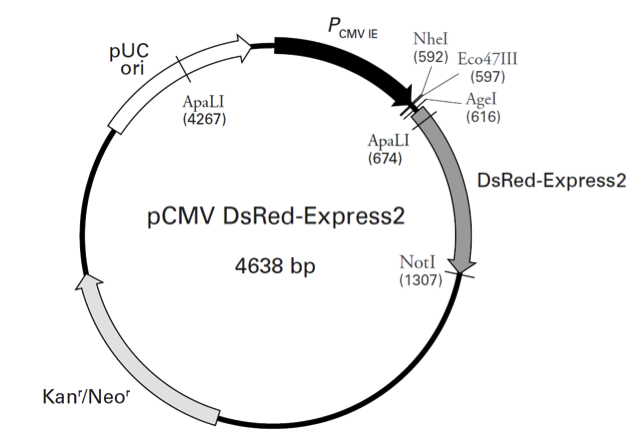
pCMV-DsRed-Express2
pCMV-DsRed-Express2
编号 | 载体名称 |
北京华越洋生物VECT6154 | pCMV-DsRed-Express2 |
pCMV-DsRed-Express2载体基本信息
载体名称: | pCMV-DsRed-Express2 |
质粒类型: | 慢病毒载体;荧光报告载体 |
高拷贝/低拷贝: | 高拷贝 |
克隆方法: | 限制性内切酶,多克隆位点 |
启动子: | CMV |
载体大小: | 4638 bp |
5' 测序引物及序列: | -- |
3' 测序引物及序列: | -- |
载体标签: | -- |
载体抗性: | 卡那霉素 |
筛选标记: | 新霉素(Neomycin) |
克隆菌株: | DH5α, HB101 |
宿主细胞(系): | 常规细胞系(293、CV-1、CHO等) |
备注: | pCMV-DsRed-Express2组成型表达DsRed-Express2; |
稳定性: | 稳表达 或 瞬表达 |
组成型/诱导型: | 组成型 |
病毒/非病毒: | 非病毒 |
pCMV-DsRed-Express2载体质粒图谱和多克隆位点信息
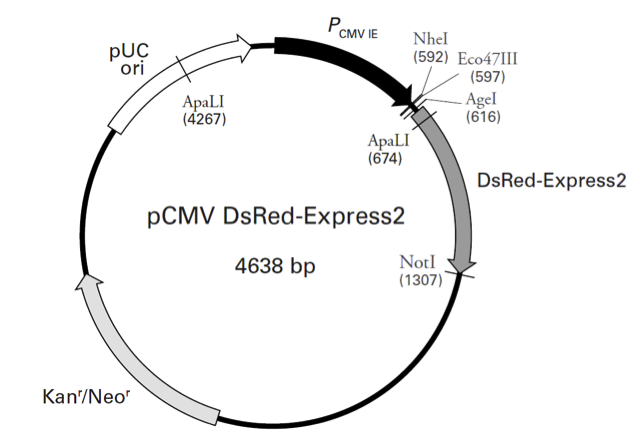

pCMV-DsRed-Express2载体描述
pCMV DsRed-Express2 is a mammalian expression vector designed to be used for whole cell labeling or as a marker of cotransfection. In mammalian cells, the vector constitutively expresses DsRed-Express2, a variant of the Discosoma sp. red fluorescent protein, DsRed (1). DsRed-Express2 retains the fast maturation and high photostability characteristic of its predecessor, DsRed-Express (2), and has been engineered (through additional amino acid substitutions) for increased solubility (3). Although it most likely forms the same tetrameric structure as wild-type DsRed, DsRed-Express2 displays a greatly reduced tendency to aggregate, resulting in reduced cyto- and phototoxicity, and making DsRed-Express2 much better suited for in vivo applications involving sensitive cells, such as primary or stem cells. In fact, DsRed-Express2 has been shown to be the best red fluorescent protein for whole cell labeling applications (3). DsRed-Express2 also exhibits extremely low residual green fluorescence, which allows cells expressing the protein to be effectively separated from other fluorescently labeled cell populations by flow cytometry. The DsRed-Express2 gene is positioned just downstream of the constitutively active human cytomegalovirus immediate early promoter (PCMV IE). As a result, mammalian cells transfected with this vector will constitutively express the red fluorescent protein. A Kozak consensus sequence (4) has been placed immediately upstream of the DsRed-Express2 coding sequence to enhance translational efficiency in eukaryotic cells. SV40 polyadenylation signals downstream of the DsRed-Express2 gene direct proper processing of the 3' end of the DsRed-Express2 mRNA. The vector backbone contains an SV40 origin for replication in mammalian cells expressing the SV40 large T antigen, a pUC origin of replication for propagation in E. coli, and an f1 origin for single-stranded DNA production. A neomycin resistance cassette (Neor) allows stably transfected eukaryotic cells to be selected using G418 (5). This cassette consists of the SV40 early promoter, a Tn5 kanamycin/neomycin resistance gene, and herpes simplex virus thymidine kinase (HSV TK) polyadenylation signals. A bacterial promoter upstream of the cassette expresses kanamycin resistance in E. coli.
pCMV DsRed-Express2 can be used for whole cell labeling or as a cotransfection marker. The vector can be transfected into mammalian cells using any standard transfection method. Cells expressing DsRed-Express2 (excitation and emission maxima: 554 nm and 591 nm, respectively) can be detected by fluorescence microscopy or flow cytometry 8-12 hours after transfection. If required, stable transfectants can be selected using G418. After cotransfection with pCMV DsRed-Express2 and an expression construct of interest, cells can also be sorted by flow cytometry to enrich for transfected cells.
Propagation in E. coli
Recommended host strain: DH5α, HB101, and other general purpose strains. Single-stranded DNA production
requires a host containing an F plasmid such as JM109 or XL1-Blue.
Selectable marker: plasmid confers resistance to kanamycin (50 μg/ml) in E. coli hosts.
E. coli replication origin: pUC
Copy number: high
Plasmid incompatibility group: pMB1/ColE1
Excitation and emission maxima of DsRed-Express2
Excitation maximum = 554 nm
Emission maximum = 591 nm
References
1. Matz, M. V. et al. (1999) Nat. Biotechnol. 17(10):969-973.
2. Bevis, B. J. & Glick, B. S. (2002) Nat. Biotechnol. 20(1):83–87. Erratum in Nat. Biotechnol. (2002) 20(11):1159
3. Strack, R. L. et al. (2008) Nat. Methods 5(11):955–957.
4. Kozak, M. (1987) Nucleic Acids Res. 15(20): 8125-8148
5. Gorman, C. (1985) In DNA Cloning: A Practical Approach, Vol. II. Ed. D. M. Glover. (IRL Press, Oxford, U.K.), pp. 143–190.
pCMV-DsRed-Express2载体序列
ORIGIN
1 TAGTTATTAA TAGTAATCAA TTACGGGGTC ATTAGTTCAT AGCCCATATA TGGAGTTCCG
61 CGTTACATAA CTTACGGTAA ATGGCCCGCC TGGCTGACCG CCCAACGACC CCCGCCCATT
121 GACGTCAATA ATGACGTATG TTCCCATAGT AACGCCAATA GGGACTTTCC ATTGACGTCA
181 ATGGGTGGAG TATTTACGGT AAACTGCCCA CTTGGCAGTA CATCAAGTGT ATCATATGCC
241 AAGTACGCCC CCTATTGACG TCAATGACGG TAAATGGCCC GCCTGGCATT ATGCCCAGTA
301 CATGACCTTA TGGGACTTTC CTACTTGGCA GTACATCTAC GTATTAGTCA TCGCTATTAC
361 CATGGTGATG CGGTTTTGGC AGTACATCAA TGGGCGTGGA TAGCGGTTTG ACTCACGGGG
421 ATTTCCAAGT CTCCACCCCA TTGACGTCAA TGGGAGTTTG TTTTGGCACC AAAATCAACG
481 GGACTTTCCA AAATGTCGTA ACAACTCCGC CCCATTGACG CAAATGGGCG GTAGGCGTGT
541 ACGGTGGGAG GTCTATATAA GCAGAGCTGG TTTAGTGAAC CGTCAGATCC GCTAGCGCTA
601 CCGGACTCAG ATCCACCGGT CGCCACCATG GATAGCACTG AGAACGTCAT CAAGCCCTTC
661 ATGCGCTTCA AGGTGCACAT GGAGGGCTCC GTGAACGGCC ACGAGTTCGA GATCGAGGGC
721 GAGGGCGAGG GCAAGCCCTA CGAGGGCACC CAGACCGCCA AGCTGCAGGT GACCAAGGGC
781 GGCCCCCTGC CCTTCGCCTG GGACATCCTG TCCCCCCAGT TCCAGTACGG CTCCAAGGTG
841 TACGTGAAGC ACCCCGCCGA CATCCCCGAC TACAAGAAGC TGTCCTTCCC CGAGGGCTTC
901 AAGTGGGAGC GCGTGATGAA CTTCGAGGAC GGCGGCGTGG TGACCGTGAC CCAGGACTCC
961 TCCCTGCAGG ACGGCACCTT CATCTACCAC GTGAAGTTCA TCGGCGTGAA CTTCCCCTCC
1021 GACGGCCCCG TAATGCAGAA GAAGACTCTG GGCTGGGAGC CCTCCACCGA GCGCCTGTAC
1081 CCCCGCGACG GCGTGCTGAA GGGCGAGATC CACAAGGCGC TGAAGCTGAA GGGCGGCGGC
1141 CACTACCTGG TGGAGTTCAA GTCAATCTAC ATGGCCAAGA AGCCCGTGAA GCTGCCCGGC
1201 TACTACTACG TGGACTCCAA GCTGGACATC ACCTCCCACA ACGAGGACTA CACCGTGGTG
1261 GAGCAGTACG AGCGCGCCGA GGCCCGCCAC CACCTGTTCC AGTAGCGGCC GCGACTCTAG
1321 ATCATAATCA GCCATACCAC ATTTGTAGAG GTTTTACTTG CTTTAAAAAA CCTCCCACAC
1381 CTCCCCCTGA ACCTGAAACA TAAAATGAAT GCAATTGTTG TTGTTAACTT GTTTATTGCA
1441 GCTTATAATG GTTACAAATA AAGCAATAGC ATCACAAATT TCACAAATAA AGCATTTTTT
1501 TCACTGCATT CTAGTTGTGG TTTGTCCAAA CTCATCAATG TATCTTAAGG CGTAAATTGT
1561 AAGCGTTAAT ATTTTGTTAA AATTCGCGTT AAATTTTTGT TAAATCAGCT CATTTTTTAA
1621 CCAATAGGCC GAAATCGGCA AAATCCCTTA TAAATCAAAA GAATAGACCG AGATAGGGTT
1681 GAGTGTTGTT CCAGTTTGGA ACAAGAGTCC ACTATTAAAG AACGTGGACT CCAACGTCAA
1741 AGGGCGAAAA ACCGTCTATC AGGGCGATGG CCCACTACGT GAACCATCAC CCTAATCAAG
1801 TTTTTTGGGG TCGAGGTGCC GTAAAGCACT AAATCGGAAC CCTAAAGGGA GCCCCCGATT
1861 TAGAGCTTGA CGGGGAAAGC CGGCGAACGT GGCGAGAAAG GAAGGGAAGA AAGCGAAAGG
1921 AGCGGGCGCT AGGGCGCTGG CAAGTGTAGC GGTCACGCTG CGCGTAACCA CCACACCCGC
1981 CGCGCTTAAT GCGCCGCTAC AGGGCGCGTC AGGTGGCACT TTTCGGGGAA ATGTGCGCGG
2041 AACCCCTATT TGTTTATTTT TCTAAATACA TTCAAATATG TATCCGCTCA TGAGACAATA
2101 ACCCTGATAA ATGCTTCAAT AATATTGAAA AAGGAAGAGT CCTGAGGCGG AAAGAACCAG
2161 CTGTGGAATG TGTGTCAGTT AGGGTGTGGA AAGTCCCCAG GCTCCCCAGC AGGCAGAAGT
2221 ATGCAAAGCA TGCATCTCAA TTAGTCAGCA ACCAGGTGTG GAAAGTCCCC AGGCTCCCCA
2281 GCAGGCAGAA GTATGCAAAG CATGCATCTC AATTAGTCAG CAACCATAGT CCCGCCCCTA
2341 ACTCCGCCCA TCCCGCCCCT AACTCCGCCC AGTTCCGCCC ATTCTCCGCC CCATGGCTGA
2401 CTAATTTTTT TTATTTATGC AGAGGCCGAG GCCGCCTCGG CCTCTGAGCT ATTCCAGAAG
2461 TAGTGAGGAG GCTTTTTTGG AGGCCTAGGC TTTTGCAAAG ATCGATCAAG AGACAGGATG
2521 AGGATCGTTT CGCATGATTG AACAAGATGG ATTGCACGCA GGTTCTCCGG CCGCTTGGGT
2581 GGAGAGGCTA TTCGGCTATG ACTGGGCACA ACAGACAATC GGCTGCTCTG ATGCCGCCGT
2641 GTTCCGGCTG TCAGCGCAGG GGCGCCCGGT TCTTTTTGTC AAGACCGACC TGTCCGGTGC
2701 CCTGAATGAA CTGCAAGACG AGGCAGCGCG GCTATCGTGG CTGGCCACGA CGGGCGTTCC
2761 TTGCGCAGCT GTGCTCGACG TTGTCACTGA AGCGGGAAGG GACTGGCTGC TATTGGGCGA
2821 AGTGCCGGGG CAGGATCTCC TGTCATCTCA CCTTGCTCCT GCCGAGAAAG TATCCATCAT
2881 GGCTGATGCA ATGCGGCGGC TGCATACGCT TGATCCGGCT ACCTGCCCAT TCGACCACCA
2941 AGCGAAACAT CGCATCGAGC GAGCACGTAC TCGGATGGAA GCCGGTCTTG TCGATCAGGA
3001 TGATCTGGAC GAAGAGCATC AGGGGCTCGC GCCAGCCGAA CTGTTCGCCA GGCTCAAGGC
3061 GAGCATGCCC GACGGCGAGG ATCTCGTCGT GACCCATGGC GATGCCTGCT TGCCGAATAT
3121 CATGGTGGAA AATGGCCGCT TTTCTGGATT CATCGACTGT GGCCGGCTGG GTGTGGCGGA
3181 CCGCTATCAG GACATAGCGT TGGCTACCCG TGATATTGCT GAAGAGCTTG GCGGCGAATG
3241 GGCTGACCGC TTCCTCGTGC TTTACGGTAT CGCCGCTCCC GATTCGCAGC GCATCGCCTT
3301 CTATCGCCTT CTTGACGAGT TCTTCTGAGC GGGACTCTGG GGTTCGAAAT GACCGACCAA
3361 GCGACGCCCA ACCTGCCATC ACGAGATTTC GATTCCACCG CCGCCTTCTA TGAAAGGTTG
3421 GGCTTCGGAA TCGTTTTCCG GGACGCCGGC TGGATGATCC TCCAGCGCGG GGATCTCATG
3481 CTGGAGTTCT TCGCCCACCC TAGGGGGAGG CTAACTGAAA CACGGAAGGA GACAATACCG
3541 GAAGGAACCC GCGCTATGAC GGCAATAAAA AGACAGAATA AAACGCACGG TGTTGGGTCG
3601 TTTGTTCATA AACGCGGGGT TCGGTCCCAG GGCTGGCACT CTGTCGATAC CCCACCGAGA
3661 CCCCATTGGG GCCAATACGC CCGCGTTTCT TCCTTTTCCC CACCCCACCC CCCAAGTTCG
3721 GGTGAAGGCC CAGGGCTCGC AGCCAACGTC GGGGCGGCAG GCCCTGCCAT AGCCTCAGGT
3781 TACTCATATA TACTTTAGAT TGATTTAAAA CTTCATTTTT AATTTAAAAG GATCTAGGTG
3841 AAGATCCTTT TTGATAATCT CATGACCAAA ATCCCTTAAC GTGAGTTTTC GTTCCACTGA
3901 GCGTCAGACC CCGTAGAAAA GATCAAAGGA TCTTCTTGAG ATCCTTTTTT TCTGCGCGTA
3961 ATCTGCTGCT TGCAAACAAA AAAACCACCG CTACCAGCGG TGGTTTGTTT GCCGGATCAA
4021 GAGCTACCAA CTCTTTTTCC GAAGGTAACT GGCTTCAGCA GAGCGCAGAT ACCAAATACT
4081 GTTCTTCTAG TGTAGCCGTA GTTAGGCCAC CACTTCAAGA ACTCTGTAGC ACCGCCTACA
4141 TACCTCGCTC TGCTAATCCT GTTACCAGTG GCTGCTGCCA GTGGCGATAA GTCGTGTCTT
4201 ACCGGGTTGG ACTCAAGACG ATAGTTACCG GATAAGGCGC AGCGGTCGGG CTGAACGGGG
4261 GGTTCGTGCA CACAGCCCAG CTTGGAGCGA ACGACCTACA CCGAACTGAG ATACCTACAG
4321 CGTGAGCTAT GAGAAAGCGC CACGCTTCCC GAAGGGAGAA AGGCGGACAG GTATCCGGTA
4381 AGCGGCAGGG TCGGAACAGG AGAGCGCACG AGGGAGCTTC CAGGGGGAAA CGCCTGGTAT
4441 CTTTATAGTC CTGTCGGGTT TCGCCACCTC TGACTTGAGC GTCGATTTTT GTGATGCTCG
4501 TCAGGGGGGC GGAGCCTATG GAAAAACGCC AGCAACGCGG CCTTTTTACG GTTCCTGGCC
4561 TTTTGCTGGC CTTTTGCTCA CATGTTCTTT CCTGCGTTAT CCCCTGATTC TGTGGATAAC
4621 CGTATTACCG CCATGCAT
//
其他哺乳动物表达载体: